|
|
| Our group has so far reported the complete genome sequences of Corynebacterium glutamicum R, Desulfitobacterium hafniense Y51, and Clostridium kluyveri. |
| Genomic analysis of Corynebacterium glutamicum R |
| Corynebacterium glutamicum has been long used for the industrial production of amino acids and nucleic acids. By using C. glutamicum R as a host, our group is working on the production of ethanol as well as organic acids including lactic acid and succinic acids which are the major components of biodegradable plastics. The genome of C. glutamicum R was 3.3Mb in size and predicted the presence of 2990 ORFs encoding 'core' genes as well as genes for transcription factors, secretion proteins, sugar metabolisms, and dual-regulation systems which are characteristic among Corynebacterium sp. |
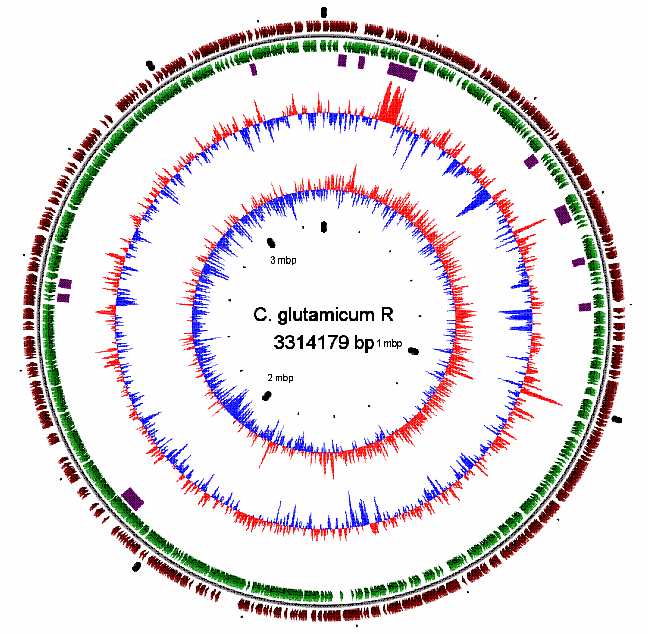 |
| C. glutamicum R genome map |
| < References > Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153: 1042-1058. 2007. H. Yukawa, C.A. Omumasaba, H. Nonaka, P. Kos, N. Okai, N. Suzuki, M. Suda, Y. Tsuge, J. Watanabe, Y. Ikeda, A.A. Vertès and M. Inui. |
| Genomic analysis of Desulfitobacterium hafniense Y51 |
| D. hafniense Y51 dehalogenates tetrachloroethylene. Sequence analysis predicted 5,060 ORFs on its 5.7 Mb-genome. Compared with other bacteria with dehalogenation capability such as D. ethenogenes 195, this bacterium possessed only two dehalogenase genes despite of its large genome size. Instead, a large number of electron acceptor/donor genes were found. |
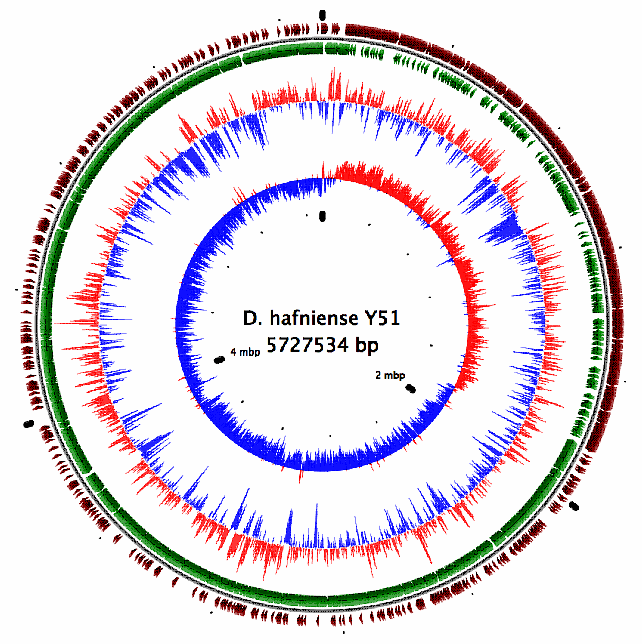 |
| D. hafniense Y51 genome map |
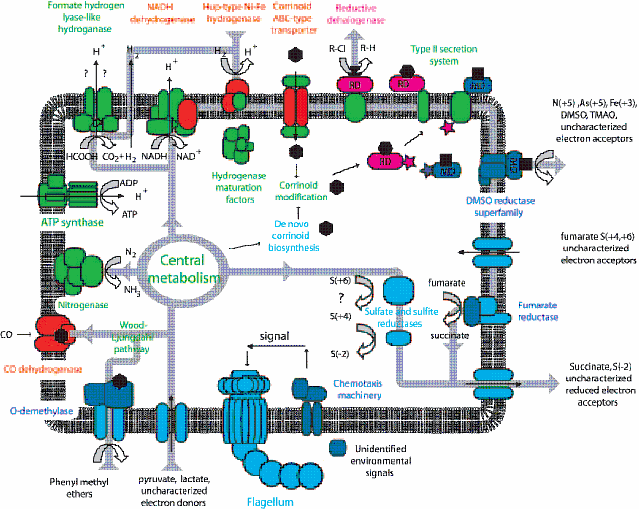 |
| D. hafniense Y51 metabolic pathway |
| < References > Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188: 2262-2274. 2006. H. Nonaka, G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui and H. Yukawa. *Genome sequence of Clostridium kluyveri (an anaerobe with a capability of producing caproic acids (C6) and butyric acids (C4) from acetic acids (C2)) will be published soon. |